4.2 Lab Exercises
Overview
This is a longer lab that takes us away from the command line.
We will do two major things in this lab:
Learn how to use the Repeatexplorer2 portal on Galaxy
Walk through the RepeatExplorer2 Nature Protocols paper and subsample shotgun data from Toomers Oak
Upload data to Galaxy
Run RepeatExplorer2 to characterize TE content in Toomers Oak
Task A
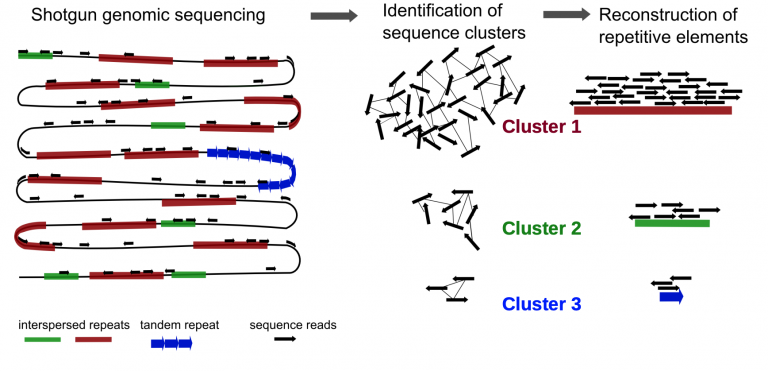
We can use Illumina paired-end shotgun genome data to predict how repetitive a genome is. Plant genomes are filled with repetitive DNA — but do we need to sequence an entire plant genome to figure out what percent of a genome is filled with repeats?
No. Imagine you have a 3 billion piece puzzle. Even if you only take 10,000 random pieces of that puzzle, you can roughly see what the puzzle should be. That premise underlies how RepeatExplorer2 works. Even with a small subsample, 1% coverage of the genome, you can nicely estimate the repetitive content of a plant genome. In short, it works by taking a small sample of reads from your genome of interest, clustering them together, and trying to reconstruct consensus sequences of repetitive elements. By doing this, you can estimate what total % of a genome is repetitive, and break down individual superfamilies of transposable elements.
Here is the main RepeatExplorer2 site to start from: https://galaxy-elixir.cerit-sc.cz/
First, Register (don’t re-use any known username or password of yours!) : https://perun.cesnet.cz/elixir2/registrar/?vo=elixir-cz&group=repeatExplorer
Then you’ll have access to the Galaxy server: https://repeatexplorer-elixir.cerit-sc.cz/galaxy/
Second, follow along with the Nature Protocols paper describing how to run RepeatExplorer2. You’ll be using the Toomers Oak PE150 PCR-free shotgun data, though. I’ve also left this pdf in the class resources for this module.